Comparison of speed, memory peak and processing results of different... | Download Scientific Diagram

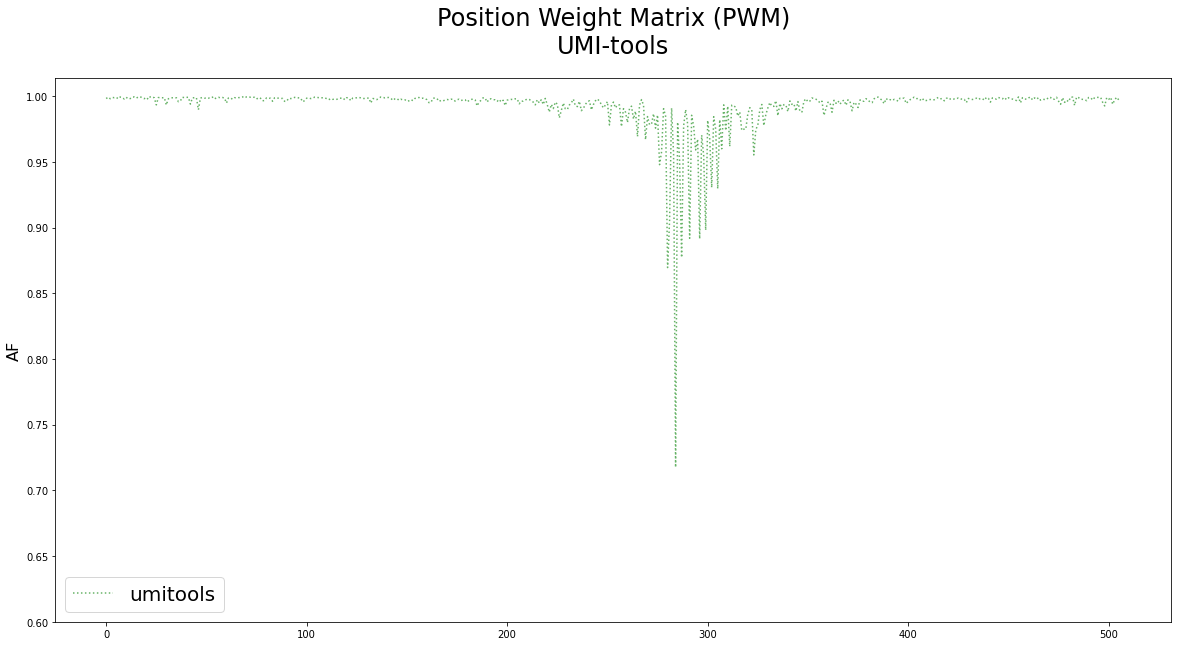
UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv

Homotrimer barcodes enable accurate counting of RNA molecules during high-throughput RNA sequencing | Nature Methods

UMI-tools: Modelling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy | bioRxiv

Analysis of UMIs. a Stacked bar plot showing the fractions of unique... | Download Scientific Diagram

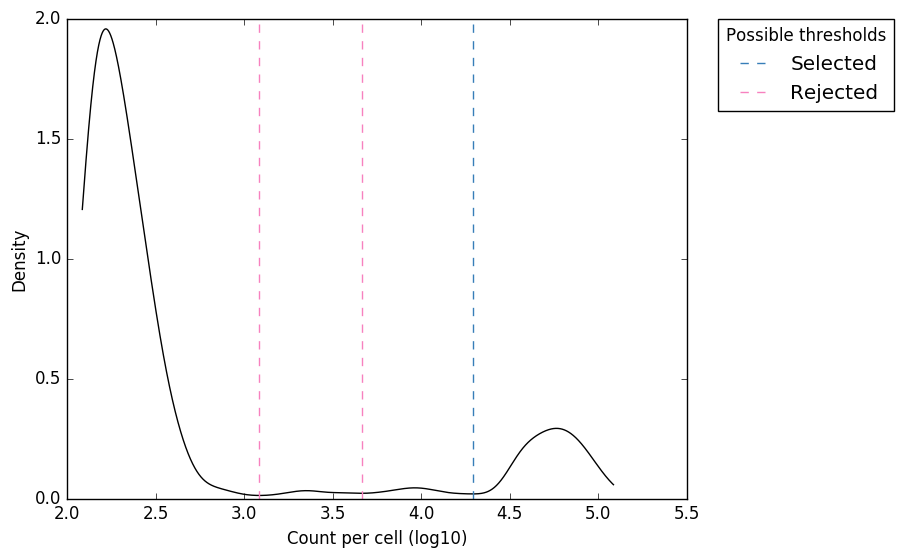
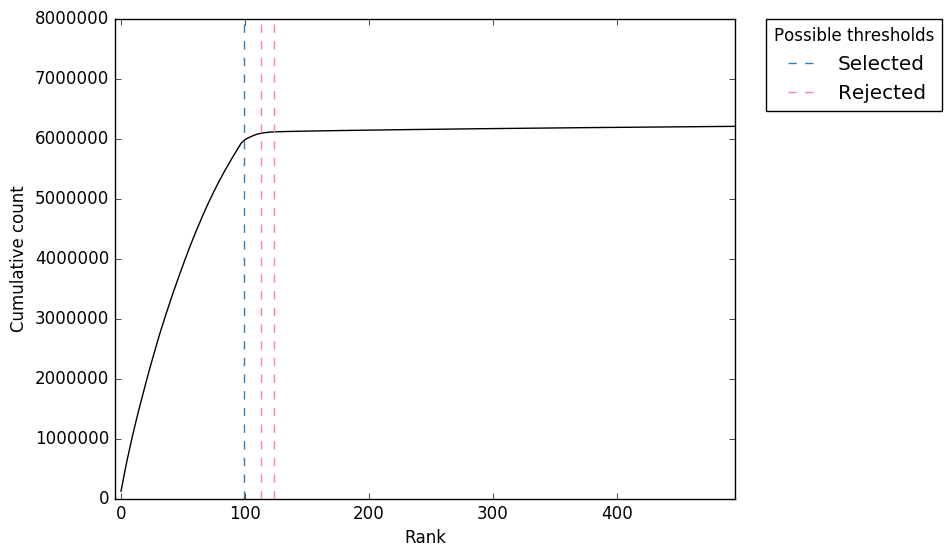
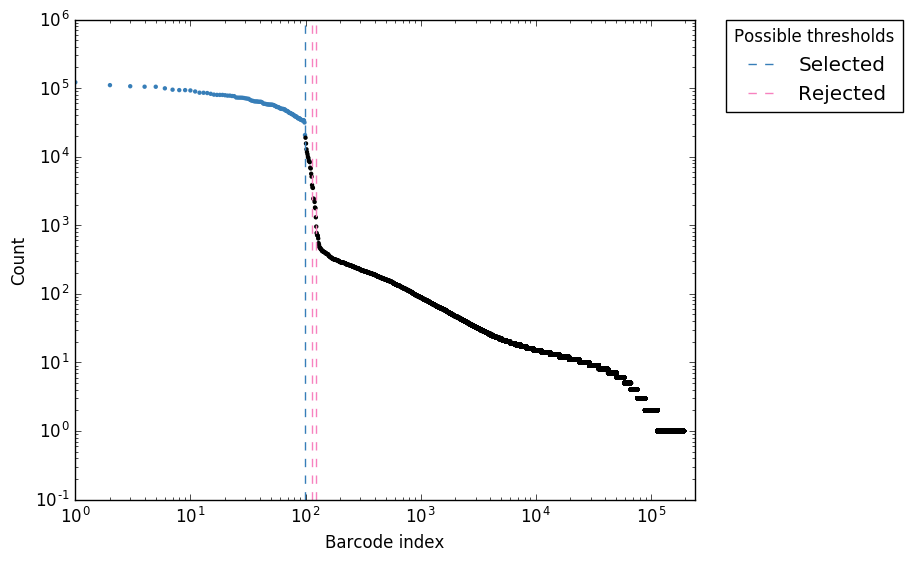
UMI-Gen: A UMI-based read simulator for variant calling evaluation in paired-end sequencing NGS libraries - ScienceDirect